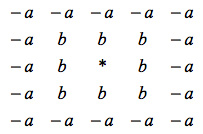Mathematical biology has always been obsessed with shapes, specially because many that look so complicated can actually be created with simple rules. Take the Turing patterns – these are regular shapes that obey the same type of rules but can make spots, stripes, labyrinths or holes by changing its parameters.

Turing-like patters in a model of biofilm, from Xavier et al (2009) Am Nat
Some years ago I read a paper by ecologists Rietkerk, Dekker, de Ruiter and van de Koppel that clicked for me. They explained that patterns in arid vegetation can have a simple explanation similar to Turing patterns. Plants in arid regions require moister (the growth limiting resource) and benefit from having other plants close by because neighbors provide shade and reduce water evaporation from the soil. However, too many neighbors means there is less moister to go around. In their words “vegetation patterns are the result of fine-scale positive feedback and coarse-scale negative feedback.” This means that having a few neighbors close by is good, but too many neighbors is bad. They presented a very simple cellular automata model that could reproduce the patterns. The cellular automaton is like a simple iterative game where the elements on a square grid follow a simple set of rules at every turn. Every element in the cellular automaton does this:
- count the number of close neighbors and multiply that number by a positive value b, which produces the total benefit of having close neighbors provide shade (B)
- count the number of neighbors over a wider range and multiply by a negative value –a, which produces the total cost of having neighbors that compete for water (-C)
In the supporting material for their paper they show a convolution kernel:
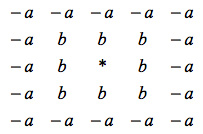
The kernel is an intuitive way to think about the cellular automaton rule. A focal bush located at the center of the kernel (represented by *) “looks” at the space around it and adds the value “b” or “-a” depending on whether that patch of land is occupied by another bush. If the final result is greater or equal to 0 then the focal bush gets to live and play another round. If the value is 0 then the bush dies and that grid element becomes free. The model is explained in detail the supporting material of the paper by Rietkerk et al. But the really striking result is that this very simple model produces a diversity of shapes depending on the value of a and b. The patterns can be made larger by using a larger kernel and also by changing the relative sizes of the “good” and “bad” neighborhoods.
We recently applied a similar idea to explain branching patterns of swarming colonies. Branching is a different process, we think, because it happens as a population spreads in space, but the patterns may have an equally simple explanation. In our case, a colony of Pseudomonas aeruginosa spreads across a petri dish and branches along the way suggesting that having too many neighbors is bad. We did some experiments that show support for these rules. For example, if we put two colonies in the same petri dish they repel each other suggesting the long range negative feedback (see video). Our model and experiments show that like the patterns in arid vegetation the branching in swarming can be caused by a short range positive feedback and a long range negative feedback (see video of model SIMSWARM).
Read the paper
The ecological basis of morphogenesis: branching patterns in swarming colonies of bacteria
Deng P, de Vargas Roditi L, van Ditmarsch D, Xavier JB. New Journal of Physics [Article: open access]
The paper has been selected to appear in the New Journal of Physics “Highlights of 2014” collection and recommended by Rob Palmer at the Faculty of 1000